Introduction
Chronic myeloid leukemia is a myeloproliferative neoplasm which develops due to the reciprocal translocation of t(22;9) forming Philadelphia chromosome, BCR::ABL fusion oncogene (1). This led to the discovery of drugs known as tyrosine kinase inhibitors (TKIs). These inhibitors bind to the kinase domain (KD) of the BCR::ABL1 oncoprotein in a competitive manner to inhibit it.(2). Nevertheless, as the disease progresses, TKIs become less effective. Unfortunately, the mechanism of CML advancement is not well known, and there are no readily available common biomarkers for CML progression (3). There are many transcription factors (TFs) which are involved in several cancers and their progression. Though the TFs are found in the progression of acute leukemias however the TFs which are exclusively associated with progression of CML are not well studied. Therefore, objective of this study was to find about novel TFs associated with CML progression.
Materials and Methods:
Patient selection: The study subjects included accelerated phase (AP-CML) and blast crisis phase CML (BC-CML) patients (Experimental groups 1 & 2, respectively) while chronic phase treatment-naïve CML patients (CP-CML) were used as control 1, CP-CML CML long-term TKI responders (at least 3 continuous years of MMR) as control group 2 and TK-resistant CML patients as control group 3, along with healthy controls.
Collection of samples: DNA extraction, and clinical monitoring. All trial participants' peripheral blood was drawn in the amount of 10 ml. During the course of this investigation, DNA was taken and patient follow-up was conducted. According to ENL guidelines, all treatment standards were followed.
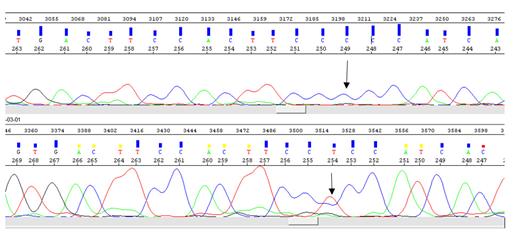
Sanger Sequencing: Sanger sequencing was carried out using genetic analyzer 3130XL. Raw reads were analyzed by using sequence analysis software and mutation surveyor.
Results:
We found MUC3A as a novel TF-gene mutated in all AP/BC-CML but in none of the controls (missense mutation 3025C>T). The original wild-type residue and newly introduced mutant residue often differ in properties (P258S). We used dbNSFP to link this mutation to a genomic variant. This variant's MetaRNN score is 0.0052194. It can range from 0.0 to 1.0. The higher, the more likely it is to be pathogenic. The mutant and wild-type residue are not very similar. Based on this conservation information this mutation is probably damaging to the protein as the mutant residue is located near a highly conserved position (4). It shows that mutated MUC3A is a novel biomarker exclusively for advanced phases of CML.
Discussion:
MUC3A belongs to the Mucin (MUC) family, a group of glycoproteins expressed in mammalian epithelial cells some of which are secreted and membrane bound, has been reported to play role in various cancers through distant signaling pathways (5-7). MUC proteins have been regarded as prospective therapeutic targets as well as biomarkers for human cancer because of their distinctive biological and structural characteristics. Hence further studies are required to find exact role of MUC3A in CML progression and its evaluation as potential biomarker and novel drug target in AP- and BC-CML.
Conflict of interest: The authors declare that they have no conflict of interests.
References:
1. Alanazi N, Iqbal Z. Blood. 2022;140(Supplement 1):12233-4.
2. Kumar V, Jyotirmayee, Verma M. Molecular and Cellular Biochemistry. 2023;478(5):1013-29.
3. Iqbal Z, Absar M, Akhtar T, Aleem A, Jameel A, Basit S, et al. Biology. 2021;10(11):1182.
4. Qin Y-M, Chen Y-Y, Liao L, Wu Y-Y, Chen M, Lin F-Q. Iranian Journal of Pediatrics. 2023(In Press).
5. Cox KE, Liu S, Lwin TM, Hoffman RM, Batra SK, Bouvet M. Cancers. 2023;15(5):1491.
6. Farhangnia P, Safdarian AR, Akbarpour M. Handbook of Cancer and Immunology: Springer; 2023. p. 1-42.
7. Yan T, Zhu L, Chen J. Experimental Hematology & Oncology. 2023;12(1):14.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal